Large study identifies biomarkers for rare kidney tumors
By Lynn McCain | May 15 2024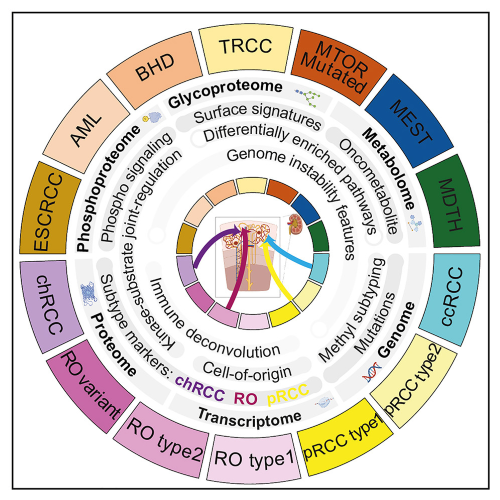 With 20 subtypes of Renal Cell Carcinoma (RCC) listed by the World Health Organization in 2022, determining the specific type of RCC and the correct treatment protocol for patients can be daunting. Only seven of the 20 subtypes had been defined by specific molecular changes and the 20% of RCCs that are classified as non-clear cell RCCs (non-ccRCC), were primarily identified only by histopathologic features. The driving genetic changes had not been identified. A large multi-institutional team making up the Clinical Proteomic Tumor Analysis Consortium, including several researchers from Michigan Medicine’s Department of Pathology, performed multi-omics data analysis on 48 non-ccRCC and 103 ccRCC tumors as well as 101 normal adjacent tissues to study the genetic aberrations found in these tumor types. The findings of this research were recently published in Cell Reports Medicine, with Pathology’s Drs. Alexey Nesvizhskii and Saravana Mohan Dhanasekaran as senior authors and Dr. Ginny Xiaohe Li as first author.
With 20 subtypes of Renal Cell Carcinoma (RCC) listed by the World Health Organization in 2022, determining the specific type of RCC and the correct treatment protocol for patients can be daunting. Only seven of the 20 subtypes had been defined by specific molecular changes and the 20% of RCCs that are classified as non-clear cell RCCs (non-ccRCC), were primarily identified only by histopathologic features. The driving genetic changes had not been identified. A large multi-institutional team making up the Clinical Proteomic Tumor Analysis Consortium, including several researchers from Michigan Medicine’s Department of Pathology, performed multi-omics data analysis on 48 non-ccRCC and 103 ccRCC tumors as well as 101 normal adjacent tissues to study the genetic aberrations found in these tumor types. The findings of this research were recently published in Cell Reports Medicine, with Pathology’s Drs. Alexey Nesvizhskii and Saravana Mohan Dhanasekaran as senior authors and Dr. Ginny Xiaohe Li as first author.
This study discovered proteogenomic, phosphorylation, glycosylation, and metabolic aberrations in RCC subtypes. RCCs with high genome instability, which are more aggressive subtypes, display overexpression of IGF2BP3 and PYCR1. The team found that RCCs develop from multiple cells-of-origin and that the various subtypes of RCC have specific proteogenomic signatures. The study improved our understanding of the biology, proteogenomics, post-translational modifications, and metabolism of kidney cancer. In addition to the IGF2BP3 and PYCR1, they identified other important biomarkers MAPRE3, ADGRF5, and GPNMB, which differentiate renal oncocytoma from chromophobe RCC, and PIGR and SOSTDC1, which distinguish papillary RCC from mucinous tubular and spindle cell carcinoma (MTSCC), a rare benign tumor.
Nesvizhskii commented, “This is a landmark study that showcases the power of integrative analyses of multi-omics data to gain a better understanding of the mechanisms that drive diseases such as cancer. Ultimately, we hope that our findings will make It possible to more accurately diagnose different subtypes of renal cancers, including rare subtypes, and will contribute to improved care for higher-risk patients.”
Citation:
Ginny Xiaohe Li, Lijun Chen, et al. Comprehensive proteogenomic characterization of rare kidney tumors. Cell Reports Medicine (2024) Published online May 3, 2024.
DOI: https://doi.org/10.1016/j.xcrm.2024.101547
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
