Study from Kleer and Nesvizhskii Labs Reveals the Proteomic Landscape of Metaplastic Breast Carcinomas
By Elizabeth Walker | May 1 2020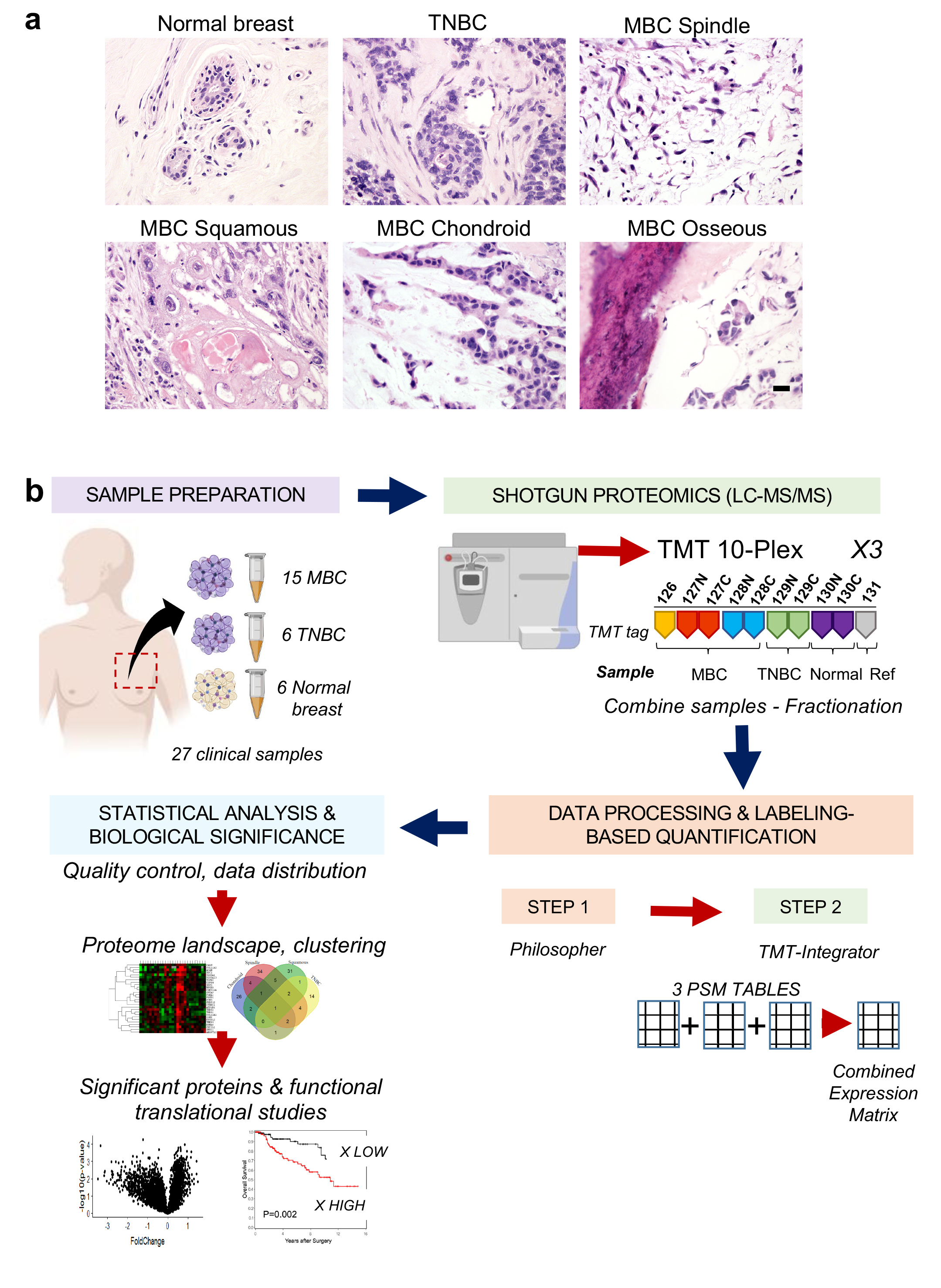 Along with co-investigators, Sabra Djomehri, a graduate student in Celina Kleer's Laboratory, has discovered the proteomic landscape of metaplastic carcinomas, the most aggressive type of triple negative breast cancer. The study, published in Nature Communications, reveals specific protein expression patterns in the different histopathological subtypes of metaplastic carcinoma. These include translational and ribosomal events in the spindle subtype, inflammation, and apical junction related proteins in squamous, and extracellular matrix proteins in sarcomatoid subtypes.
Along with co-investigators, Sabra Djomehri, a graduate student in Celina Kleer's Laboratory, has discovered the proteomic landscape of metaplastic carcinomas, the most aggressive type of triple negative breast cancer. The study, published in Nature Communications, reveals specific protein expression patterns in the different histopathological subtypes of metaplastic carcinoma. These include translational and ribosomal events in the spindle subtype, inflammation, and apical junction related proteins in squamous, and extracellular matrix proteins in sarcomatoid subtypes.
The lab is excited to provide insights into the protein profiles underlying the pathological diversity and aggressiveness of these tumors and hopes that these results will aid in the development of new therapeutics. Read the entire paper at Nature.com.
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
