
The 1990s was a time of advancements toward a future of lasting change in the world. Grunge and hip-hop were new, and cable tv came on the scene. The Hubble telescope was launched, and Dolly the sheep was the first mammal cloned from an adult somatic cell. The internet was booming as more businesses and households were purchasing personal computers, bringing in a new era of communication. And Pathology was stepping into a new frontier of what would become digital pathology. Dr. Ul Balis, Professor and Director of Pathology Informatics at Michigan Medicine, was at the forefront of this new field. Balis built a real-time telepathology linkage between the James A. Haley VA Hospital and the Bay Pines VA Hospital while in medical school, one of the first of its kind. With this, images viewed at one site could also be seen at the other, basically roboticizing a microscope. And the era of digital pathology was launched.
 Throughout the 1990s, Balis continued to work in digital pathology, sitting on the College of American Pathology Informatics Committee and forming the Image Exchange Committee, which created the first Digital Imaging and Communications in Medicine (DICOM) standard for digital pathology, the visible light standard; ratified in 1999. In 2000, he began advising Aperio, the developer of the first whole-slide imaging scanner. Through his and others’ efforts, this nascent field continued to grow and expand. On the other end of the spectrum, Dr. Liron Pantanowitz was serving as the Director of Pathology Informatics at Baystate Medical Center, one of the hospitals affiliated with Tufts University School of Medicine, when in 2005, he was asked to bring on an image analysis system for breast biomarkers. He discovered there were few resources and very limited knowledge on how to do this at the time. It required significant effort, but he successfully implemented such a system and then continued to work in the field, promoting the field of digital pathology, developing national guidelines, and engaging with organizations that educate about digital pathology.
Throughout the 1990s, Balis continued to work in digital pathology, sitting on the College of American Pathology Informatics Committee and forming the Image Exchange Committee, which created the first Digital Imaging and Communications in Medicine (DICOM) standard for digital pathology, the visible light standard; ratified in 1999. In 2000, he began advising Aperio, the developer of the first whole-slide imaging scanner. Through his and others’ efforts, this nascent field continued to grow and expand. On the other end of the spectrum, Dr. Liron Pantanowitz was serving as the Director of Pathology Informatics at Baystate Medical Center, one of the hospitals affiliated with Tufts University School of Medicine, when in 2005, he was asked to bring on an image analysis system for breast biomarkers. He discovered there were few resources and very limited knowledge on how to do this at the time. It required significant effort, but he successfully implemented such a system and then continued to work in the field, promoting the field of digital pathology, developing national guidelines, and engaging with organizations that educate about digital pathology.
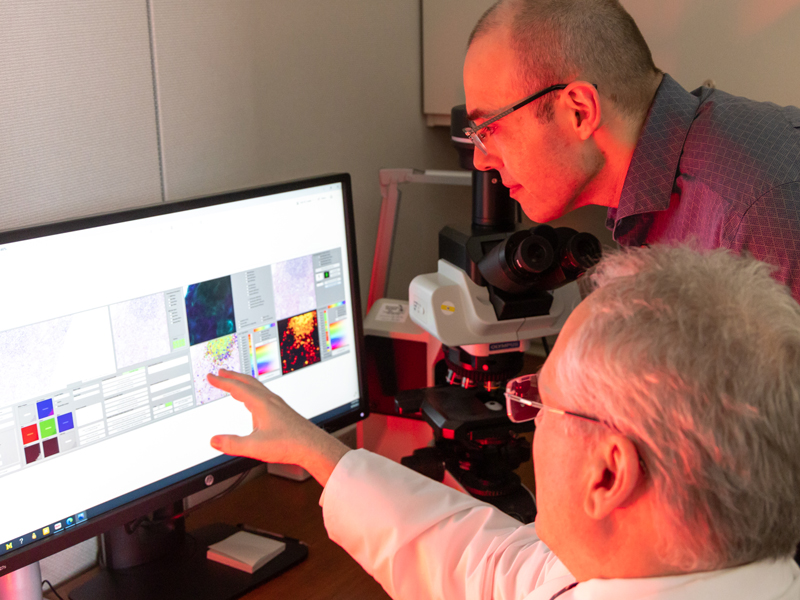
Fast forward 17 years and today there is a broad field entitled Computational Pathology that encompasses anything that has to do with computers in the field of pathology. Under this umbrella, there are whole-slide imaging systems, artificial intelligence, computer vision, as well as all the standard information systems, computer-aided laboratory equipment, and computational programs. Computer vision uses machine learning and “deep learning” to translate digitized slide images into diagnostic images using heat maps and other features to identify the areas of the original image most likely to contain abnormal cells or structures. Machine learning occurs when algorithms learn from the data you input to classify that data in a specific way. Training is supervised or semi-supervised by a subject matter expert. Deep learning, on the other hand, utilizes artificial neural networks, often hundreds of layers deep, to perform analyses and make decisions on its own. It works utilizing a completely unsupervised process, although the results are still reviewed by subject matter experts. “Deep learning requires large data sets of images, for example, and the algorithm itself decides what features distinguish classes of images and what those features are. It has the potential to extract novel classification and grading systems that pathologists don’t currently use based on an entirely novel set of visual cues or features,” explained Dr. Jerome Cheng, Associate Director of Pathology Informatics. “It is very powerful, but it is also potentially very dangerous in that it may be detecting artifacts. It has to be used with subject matter expertise as oversight.” Balis expanded, “Deep learning is certainly the newest set of tools in our toolbox, but it should not be considered the only tool. It is one of many and it works in some cases and it doesn’t work in others. It needs to be thoughtfully applied.”
"Today, we use things like stage and grade in cancer to come up with some sort of a prediction. But deep learning has the capability of an artificial brain..."
Deep learning has exciting potential applications, according to Pantanowitz. “What if we train the algorithm to do much more than a pathologist can do at the microscope? What if we want to predict who will respond to therapy and who will not? What if we wanted it to predict who will recur with a disease and who will not? Who will be alive in 5 years and who will not? Today, we use things like stage and grade in cancer to come up with some sort of prediction. But deep learning has the capability of an artificial brain, and it is exciting that Jerome and Ul can write these algorithms for us to do so much more than pathologists can do at the microscope. That is why we are investing in the technology and that is why we want to bring it on in-house – to be able to do those kinds of things.”
Deep learning is a “future state” application being developed in pathology, but in the present, the Department of Pathology continues to move forward with digital pathology applications. Digital pathology is the process of taking pathology assets, such as tissue slides, culture smears, etc. and converting them into digital images. “Then the pathologist remotely accesses and controls a computer, which in turn, controls a robotic microscope,” explained Peter Ouillette, Digital Pathology Operations Manager. “This gives the pathologist the freedom to remotely manipulate the microscopic image, look at the tissue, and focus as they see fit.” The department has used digital pathology, specifically whole-slide imaging, since 2006, primarily in education and research settings. “We have built up a public repository of whole-slide images for use by pathologists and trainees here at Michigan as well as around the world,” said Ouillette. This has enabled faculty and trainees to become comfortable with the technology. In education, residents and fellows are able to view digital slides as part of their training from any location–whether they are at home, on service at the hospital or in the resident suite at the North Campus Research Complex. The large digital slide library enables researchers to access the assets they need for their research projects, publications, and presentations without ever leaving their office or lab.
 Telepathology, which utilizes whole-slide images, robotic microscopy and/or real-time video streaming viewed remotely, has significantly expanded the ability of pathologists to support the clinical work of Michigan Medicine. A large proportion of the tumor boards now utilize this technology. Rather than having specialists from around the medical campus meet in a conference room to discuss cases, they view the cases remotely and meet virtually, which enables broader representation at tumor boards and improved patient care.
Telepathology, which utilizes whole-slide images, robotic microscopy and/or real-time video streaming viewed remotely, has significantly expanded the ability of pathologists to support the clinical work of Michigan Medicine. A large proportion of the tumor boards now utilize this technology. Rather than having specialists from around the medical campus meet in a conference room to discuss cases, they view the cases remotely and meet virtually, which enables broader representation at tumor boards and improved patient care.
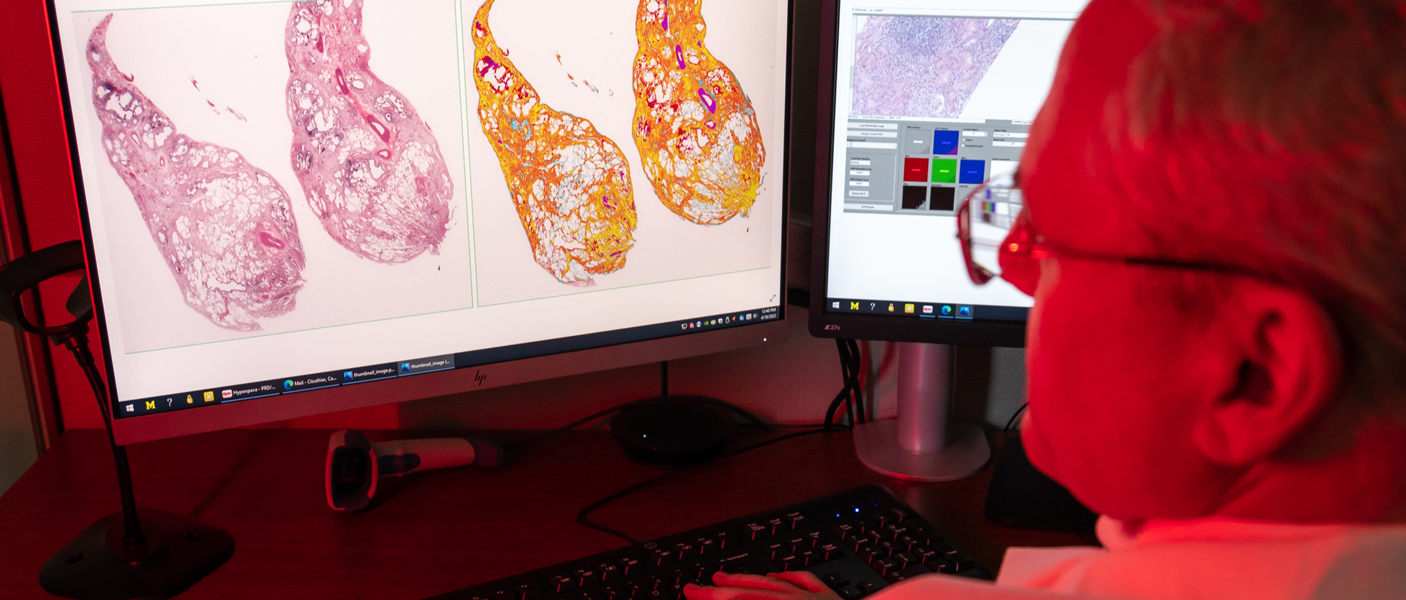
In addition to simply viewing digital images, the department is increasingly using image analysis, which uses algorithms to identify and quantify areas of interest. “In breast pathology, all our newly diagnosed breast carcinoma cases are analyzed using quantitative image analysis (QIA). Each of these cases have biomarker studies completed on them, ER, PR, and HER2/neu. QIA is also used to evaluate Ki67 (a proliferation marker) for neuroendocrine tumors. Using QIA, we now have a very standardized analysis and reporting mechanism, so it doesn’t matter which pathologist is on service. They all use the same yardstick,” explained Pantanowitz. “In fact, this is so reliable that we have outside hospitals send us their cases to report on.”
“To date, our digital efforts have all been confirmatory analyses performed after the pathologists had reviewed the slides. As much as we wanted to do so, we were unable to bring this technology to the general clinical setting for primary diagnostics due to regulatory constraints,” explained Pantanowitz. “With the onset of the pandemic, however, many of these constraints were lifted, enabling pathologists to diagnose cases remotely.” Over the course of the pandemic, the department moved from confirmatory analyses to limited primary diagnoses using digital pathology. Pathologists were having to quarantine, or they couldn’t come to the clinic due to childcare issues or other pandemic-related issues. Processes were validated and regulatory requirements, some of which were under waiver, were met. The Pathology Informatics team then provided pathologists with the technology and training needed to perform remote diagnoses for patient cases. “While everyone is now back to work, some of our pathologists don’t want to give up the convenience and flexibility of remotely utilizing digital pathology.” Pantanowitz illustrated this point, “We have one faculty member who couldn’t come to work for several weeks. He was able to continue signing out cases from home. Now that he is back to work, he doesn’t want to give that up! He loves the convenience.”
"In the future, it may be easier for certain outside healthcare providers to send us their images for consultations than to mail us slides and blocks, which expands our reach and reduces turn-around times for consultations."
Balis added, “In addition to using digital pathology for primary diagnoses, this application is ideal for intraoperative consultations and rapid onsite evaluation in cytology to ensure specimen adequacy during biopsies and fine needle aspirations. The pathologists can ensure proper samples were procured without leaving their offices. They are virtually ‘at the patient bedside’ looking at what is coming back from CT-guided or other aspiration biopsies from multiple surgical suites at the same time.” This means that patients undergoing biopsies spend less time in procedure rooms awaiting pathologist consultation completion. It can take 20-30 minutes for a pathologist to walk from the University Hospital to the Mott Children’s and Women’s Hospital to perform these frozen section tissue reviews. In addition to the hospital surgical suites, there are also several located at outpatient sites. It is not feasible to keep each site fully staffed with neuropathologists, frozen section support, cytopathologists, and other services on a daily basis. “This has allowed Michigan Medicine to open up endocrine clinics and still do thyroid FNAs with onsite evaluation in convenient facilities around Michigan Medicine,” said Pantanowitz. “In the future, it may be easier for certain outside healthcare providers to send us their images for consultations than to mail us slides and blocks, which expands our reach and reduces turn-around times for consultations.” In addition to the surgical reviews, the microbiology laboratory has found digital pathology to be particularly useful. Whole-slide imaging of smears and stains of bacteria, parasites, and fungal organisms are viewed and diagnoses made quickly, which can allow patients to receive the appropriate antimicrobial therapy sooner, which leads to better patient outcomes.
Beyond standard digital pathology is a newer application utilizing deep learning to provide image analysis on slides prior to pathologist review, which streamlines workloads for pathologists. Cheng is actively developing these applications for use in pathology. “The pathologist is actively directed to what needs to be reviewed, where the highest value locations are on the slide. This is more efficient, and pathologists are less likely to miss major findings because the slide has already been pre-screened. It is like having a digital resident or fellow.”
While the promise of digital pathology is exciting, there are some very significant barriers slowing its adoption. First, training the algorithms used in deep learning applications is extremely time-consuming and requires very large data sets. This is unreimbursed time, which is costly. In addition, the data storage and processing needs are intense and require large, high-powered computing clusters, either in-house or in cloud-based servers, both of which can make any positive return on investment nearly impossible, especially since insurance coverage for digital pathology is minimal. These barriers need to be examined and overcome before digital pathology will live up to its promise. Other barriers include regulatory constraints. While the lab itself is not regulated by the FDA, some of the equipment is, which limits which equipment can be utilized.
In addition, the Clinical Laboratory Improvement Act (CLIA) stipulates lab operations must take place within a CLIA-certified facility. While this is currently waived for digital pathology, if the waiver is lifted, it will prevent digital pathology from reaching its potential. Finally, laboratory information systems often don’t integrate well with digital pathology systems, which can make it challenging to implement. There are no pathology informatics interoperability standards, which need to be developed for digital pathology to reach its true potential.
"While this technology is not yet being utilized in pathology at Michigan Medicine, it is on the roadmap."
In spite of these barriers, the field continues to advance. One exciting opportunity is use of 3-D images of organs for diagnostic purposes. 3-D imaging is already in use in other medical fields, such as radiology, and in very limited use in a few pathology settings. While this technology is not yet being utilized in pathology at Michigan Medicine, it is on the roadmap. 3-D imaging is an excellent option for Anatomic Pathology’s gross examinations, where shape, color, and texture are used to determine disease state or cause of death in forensic applications. These images can be used for education, research, courtroom testimony, as well as in making diagnostic findings. The images can be viewed in 2-D, as on a computer screen, or in 3-D using virtual reality appIications. In addition, 3-D imaging can be used with 3-D printers to produce replicas that can be viewed and handled. These printers are now capable of creating life-like specimens using as many as 10 million colors for realistic representations. Transplant patients, in particular, appreciate the ability to view and handle explanted organs. Currently, “plasticized” organs can be used to educate patients. This is an expensive process, however, so patients are generally able to view only representative organs, not their own. The 3-D models allow for safe handling of otherwise biohazardous specimens and patients will be able to physically see accurate replicas of their own explanted organs.
Another advance is the use of light-sheet microscopy for slide-free, non-destructive ex-vivo microscopy (EVM). Light-sheet microscopy is a fluorescence microscopy technique with unparalleled ability to rapidly collect 3-D microscopic information from intact specimens. Open-top light sheet microscopic imaging is particularly well-suited for use in Pathology. The advantage of this method is that the tissue can be examined at multiple depths, which can provide a more complete diagnostic picture than a fixed slide image seen under a microscope, while maintaining a very similar look to the actual slide images.
Finally, “in vivo imaging” – imaging done in the patient without ever making a slide, is advancing. Dr. Sandra Camelo-Piragua, a neuropathologist in our department, has been working with neurosurgeons to image brain tissue during surgery. “They use magnetic resonance spectroscopy, and she helps them interpret the images in the patient. Right now, tissue is still being submitted for correlation, but this may eventually eliminate the need for tissue,” explained Balis.
The field of pathology has come a long way since the 1990s, and more exciting advances are on the horizon. The Department of Pathology at Michigan Medicine has the expertise to continue to move the bar forward in both creation of new advances as well as in adoption of new applications. It is an exciting time in pathology. Stay tuned…there is more to come!